Kozak Consensus Sequence on:
[Wikipedia]
[Google]
[Amazon]
The Kozak consensus sequence (Kozak consensus or Kozak sequence) is a nucleic acid motif that functions as the
NC-IUB, 1984. # 'R' indicates that a
 The scanning mechanism of Initiation starts when the PIC binds the 5′ end of the mRNA. Scanning is stimulated by
The scanning mechanism of Initiation starts when the PIC binds the 5′ end of the mRNA. Scanning is stimulated by
Lmx1b
is an example of a gene with a weak Kozak consensus sequence. For initiation of translation from such a site, other features are required in the mRNA sequence in order for the ribosome to recognize the initiation codon. Exceptions to the first AUG rule may occur if it is not contained in a Kozak-like sequence. This is called
 Research has shown that a mutation of G—>C in the −6 position of the β-globin gene (β+45; human) disrupted the haematological and biosynthetic phenotype function. This was the first mutation found in the Kozak sequence and showed a 30% decrease in translational efficiency. It was found in a family from the Southeast Italy and they suffered from thalassaemia intermedia.
Similar observations were made regarding mutations in the position −5 from the start codon, AUG. Cytosine in this position, as opposed to thymine, showed more efficient translation and increased expression of the platelet adhesion receptor, glycoprotein Ibα in humans.
Mutations to the Kozak sequence can also have drastic effects upon human health, in particular the heart disease with the
Research has shown that a mutation of G—>C in the −6 position of the β-globin gene (β+45; human) disrupted the haematological and biosynthetic phenotype function. This was the first mutation found in the Kozak sequence and showed a 30% decrease in translational efficiency. It was found in a family from the Southeast Italy and they suffered from thalassaemia intermedia.
Similar observations were made regarding mutations in the position −5 from the start codon, AUG. Cytosine in this position, as opposed to thymine, showed more efficient translation and increased expression of the platelet adhesion receptor, glycoprotein Ibα in humans.
Mutations to the Kozak sequence can also have drastic effects upon human health, in particular the heart disease with the
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
initiation site in most eukaryotic
Eukaryotes () are organisms whose Cell (biology), cells have a cell nucleus, nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the ...
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
transcripts. Regarded as the optimum sequence for initiating translation in eukaryotes, the sequence is an integral aspect of protein regulation and overall cellular health as well as having implications in human disease. It ensures that a protein is correctly translated from the genetic message, mediating ribosome assembly and translation initiation. A wrong start site can result in non-functional proteins. As it has become more studied, expansions of the nucleotide sequence, bases of importance, and notable exceptions have arisen. The sequence was named after the scientist who discovered it, Marilyn Kozak
Marilyn S. Kozak is an American professor of biochemistry at the Robert Wood Johnson Medical School. She was previously at the University of Medicine and Dentistry of New Jersey before the school was merged. She was awarded a PhD in microbiolog ...
. Kozak discovered the sequence through a detailed analysis of DNA genomic sequences.
The Kozak sequence is not to be confused with the ribosomal binding site A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers t ...
(RBS), that being either the 5′ cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation o ...
of a messenger RNA or an internal ribosome entry site An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. In eukaryotic translation, initiation typically occurs at ...
(IRES).
Sequence
The Kozak sequence was determined by sequencing of 699 vertebrate mRNAs and verified bysite-directed mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesi ...
. While initially limited to a subset of vertebrates (''i.e.'' human, cow, cat, dog, chicken, guinea pig, hamster, mouse, pig, rabbit, sheep, and ''Xenopus
''Xenopus'' () (Gk., ξενος, ''xenos''=strange, πους, ''pous''=foot, commonly known as the clawed frog) is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described within it. The two best-know ...
''), subsequent studies confirmed its conservation in higher eukaryotes generally. The sequence was defined as 5'-(gcc)gccRccAUGG-3' (IUPAC nucleobase
Nucleobases, also known as ''nitrogenous bases'' or often simply ''bases'', are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic b ...
notation summarized here) where:
# The underlined nucleotides indicate the translation start codon, coding for Methionine.
# upper-case letters indicate highly conserved bases, ''i.e.'' the 'AUGG' sequence is constant or rarely, if ever, changes.Nomenclature for Incompletely Specified Bases in Nucleic Acid SequencesNC-IUB, 1984. # 'R' indicates that a
purine
Purine is a heterocyclic aromatic organic compound that consists of two rings ( pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines ...
(adenine
Adenine () ( symbol A or Ade) is a nucleobase (a purine derivative). It is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The three others are guanine, cytosine and thymine. Its deri ...
or guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is c ...
) is always observed at this position (with adenine
Adenine () ( symbol A or Ade) is a nucleobase (a purine derivative). It is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The three others are guanine, cytosine and thymine. Its deri ...
being more frequent according to Kozak)
# a lower-case letter denotes the most common base at a position where the base can nevertheless vary
# the sequence in parentheses (gcc) is of uncertain significance.
The AUG is the initiation codon encoding a methionine amino acid at the N-terminus of the protein. (Rarely, GUG is used as an initiation codon, but methionine is still the first amino acid as it is the met-tRNA in the initiation complex that binds to the mRNA). Variation within the Kozak sequence alters the "strength" thereof. Kozak sequence strength refers to the favorability of initiation, affecting how much protein is synthesized from a given mRNA. The A nucleotide of the "AUG" is delineated as +1 in mRNA sequences with the preceding base being labeled as −1. For a 'strong' consensus, the nucleotides at positions +4 (i.e. G in the consensus) and −3 (i.e. either A or G in the consensus) relative to the +1 nucleotide must both match the consensus (there is no 0 position). An 'adequate' consensus has only 1 of these sites, while a 'weak' consensus has neither. The cc at −1 and −2 are not as conserved, but contribute to the overall strength. There is also evidence that a G in the -6 position is important in the initiation of translation. While the +4 and the −3 positions in the Kozak sequence have the greatest relative importance in the establishing a favorable initiation context a CC or AA motif at −2 and −1 were found to be important in the initiation of translation in tobacco and maize plants. Protein synthesis in yeast was found to be highly affected by composition of the Kozak sequence in yeast, with adenine enrichment resulting in higher levels of gene expression. A suboptimal Kozak sequence can allow for PIC to scan past the first AUG site and start initiation at a downstream AUG codon.
Ribosome assembly
The ribosome assembles on the start codon (AUG), located within the Kozak sequence. Prior to translation initiation, scanning is done by the pre-initiation complex (PIC). The PIC consists of the 40S (small ribosomal subunit) bound to the ternary complex,eIF2
Eukaryotic Initiation Factor 2 (eIF2) is an eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer c ...
-GTP-intiatorMet tRNA (TC) to form the 43S ribosome. Assisted by several other initiation factors ( eIF1 and eIF1A, eIF5
Eukaryotic translation initiation factor 5 is a protein that in humans is encoded by the ''EIF5'' gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ...
, eIF3
Eukaryotic initiation factor 3 (eIF3) is a multiprotein complex that functions during the initiation phase of eukaryotic translation. It is essential for most forms of cap-dependent and cap-independent translation initiation. In humans, eIF3 co ...
, polyA binding protein) it is recruited to the 5′ end of the mRNA. Eukaryotic mRNA is capped with a 7-methylguanosine (m7G) nucleotide which can help recruit the PIC to the mRNA and initiate scanning. This recruitment to the m7G 5′ cap is supported by the inability of eukaryotic ribosomes to translate circular mRNA, which has no 5′ end. Once the PIC binds to the mRNA it scans until it reaches the first AUG codon in a Kozak sequence. This scanning is referred to as the scanning mechanism of initiation.
 The scanning mechanism of Initiation starts when the PIC binds the 5′ end of the mRNA. Scanning is stimulated by
The scanning mechanism of Initiation starts when the PIC binds the 5′ end of the mRNA. Scanning is stimulated by Dhx29
DExH-box helicase 29 (DHX29) is a 155 kDa protein that in humans is encoded by the DHX29 gene.
Function
This gene encodes a member of the DEAH (Asp-Glu-Ala-His) subfamily of proteins, part of the DEAD (Asp-Glu-Ala-Asp) box family of RNA helic ...
and Ddx3/Ded1 and eIF4 proteins. The Dhx29 and Ddx3/Ded1 are DEAD-box helicases that help to unwind any secondary mRNA structure which could hinder scanning. The scanning of an mRNA continues until the first AUG codon on the mRNA is reached, this is known as the "First AUG Rule". While exceptions to the "First AUG Rule" exist, most exceptions take place at a second AUG codon that is located 3 to 5 nucleotides downstream from the first AUG, or within 10 nucleotides from the 5′ end of the mRNA. At the AUG codon a Methionine tRNA anticodon is recognized by mRNA codon. Upon base pairing to the start codon the eIF5
Eukaryotic translation initiation factor 5 is a protein that in humans is encoded by the ''EIF5'' gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ...
in the PIC helps to hydrolyze a guanosine triphosphate (GTP) bound to the eIF2. This leads to the a structural rearrangement that commits the PIC to binding to the large ribosomal subunit (60S) and forming the ribosomal complex (80S). Once the 80S ribosome complex is formed then the elongation phase of translation starts.
The first start codon closest to the 5′ end of the strand is not always recognized if it is not contained in a Kozak-like sequenceLmx1b
is an example of a gene with a weak Kozak consensus sequence. For initiation of translation from such a site, other features are required in the mRNA sequence in order for the ribosome to recognize the initiation codon. Exceptions to the first AUG rule may occur if it is not contained in a Kozak-like sequence. This is called
leaky scanning
Leaky scanning is a mechanism used during the initiation phase of eukaryotic translation that enables regulation of gene expression. During initiation, the small 40S ribosomal subunit (as a 43S PIC) "scans" or moves in a 5' --> 3' direction along ...
and could be a potential way to control translation through initiation. For initiation of translation from such a site, other features are required in the mRNA sequence in order for the ribosome to recognize the initiation codon.
It is believed that the PIC is stalled at the Kozak sequence by interactions between eIF2 and the −3 and +4 nucleotides in the Kozak position. This stalling allows the start codon and the corresponding anticodon time to form the correct hydrogen bonding. The Kozak consensus sequence is so common that the similarity of the sequence around the AUG codon to the Kozak Sequence is used as a criterion for finding start codons in eukaryotes.
Differences from bacterial initiation
The scanning mechanism of initiation, which utilizes the Kozak sequence, is found only in eukaryotes and has significant differences from the way bacteria initiate translation. The biggest difference is the existence of the Shine-Dalgarno (SD) sequence in mRNA for bacteria. The SD sequence is located near the start codon which is in contrast to the Kozak sequence which actually contains the start codon. The Shine Dalgarno sequence allows the 16S subunit of the small ribosome subunit to bind to the AUG start codon immediately with no need for scanning along the mRNA. This results in a more rigorous selection process for the AUG codon than in bacteria. An example of bacterial start codon promiscuity can be seen in the uses alternate start codons UUG and GUG for some genes. Archaeal transcripts use a mix of SD sequence, Kozak sequence, and leaderless initiation. Haloarchaea are known to have a variant of the Kozak consensus sequence in theirHsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an import ...
genes.
Mutations and disease
Marilyn Kozak demonstrated, through systematic study of point mutations, that any mutations to a strong consensus sequence in the −3 position or to the +4 position resulted in highly impaired translation initiation both ''in vitro'' and ''in vivo''. Research has shown that a mutation of G—>C in the −6 position of the β-globin gene (β+45; human) disrupted the haematological and biosynthetic phenotype function. This was the first mutation found in the Kozak sequence and showed a 30% decrease in translational efficiency. It was found in a family from the Southeast Italy and they suffered from thalassaemia intermedia.
Similar observations were made regarding mutations in the position −5 from the start codon, AUG. Cytosine in this position, as opposed to thymine, showed more efficient translation and increased expression of the platelet adhesion receptor, glycoprotein Ibα in humans.
Mutations to the Kozak sequence can also have drastic effects upon human health, in particular the heart disease with the
Research has shown that a mutation of G—>C in the −6 position of the β-globin gene (β+45; human) disrupted the haematological and biosynthetic phenotype function. This was the first mutation found in the Kozak sequence and showed a 30% decrease in translational efficiency. It was found in a family from the Southeast Italy and they suffered from thalassaemia intermedia.
Similar observations were made regarding mutations in the position −5 from the start codon, AUG. Cytosine in this position, as opposed to thymine, showed more efficient translation and increased expression of the platelet adhesion receptor, glycoprotein Ibα in humans.
Mutations to the Kozak sequence can also have drastic effects upon human health, in particular the heart disease with the GATA4
Transcription factor GATA-4 is a protein that in humans is encoded by the ''GATA4'' gene.
Function
This gene encodes a member of the GATA family of zinc finger transcription factors. Members of this family recognize the GATA motif which is pr ...
gene. The GATA4 gene is responsible for gene expression in a wide variety of tissues including the heart. When the guanosine at the -6 position in the Kozak sequence of GATA4 is mutated to a cytosine a reduction in GATA4 protein levels, which leads atrial septal defect in the heart.
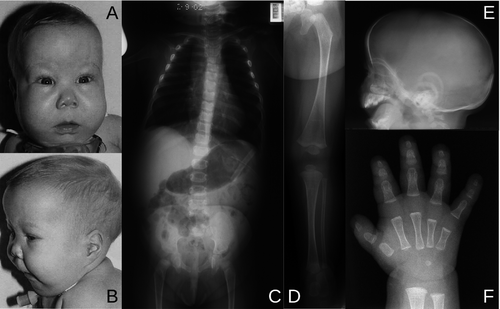
The ability of the Kozak sequence to start translation can result in novel initiation codons in the typically untranslated region of the 5′ (5′ UTR) end of the mRNA transcript. When a G to A mutation was observed in this region it resulted in an out of frame and thus protein mutation. This mutated protein results in campomelic dysplasia. Campomelic dysplasia is a developmental disorder that results in skeletal malformations.
Variations in the consensus sequence
The Kozak consensus has been variously described as: 65432-+234 (gcc)gccRccAUGG (Kozak 1987) AGNNAUGN ANNAUGG ACCAUGG (Spotts et al., 1997, mentioned in Kozak 2002) GACACCAUGG (''H. sapiens HBB, HBD'', ''R. norvegicus Hbb'', etc.)See also
*mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
, the nucleic acid messenger that serves as the middleman in the Central Dogma of Biology
* Ribosome, the molecular machine responsible for protein synthesis
* Shine–Dalgarno sequence
The Shine–Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (mRNA) to in ...
, the ribosomal binding site of prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Conne ...
s.
* Translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
, the process of peptide synthesis
References
Further reading
* * * {{GeneticTranslation Protein biosynthesis